Visualization
import numpy as np
import pandas as pd
from sklearn.cross_validation import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_curve, auc
import matplotlib as mpl
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import pandas as pd
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
pd.set_option('display.notebook_repr_html', True)
import seaborn as sns
sns.set_style("whitegrid")
sns.set_context("poster")
from matplotlib.colors import ListedColormap
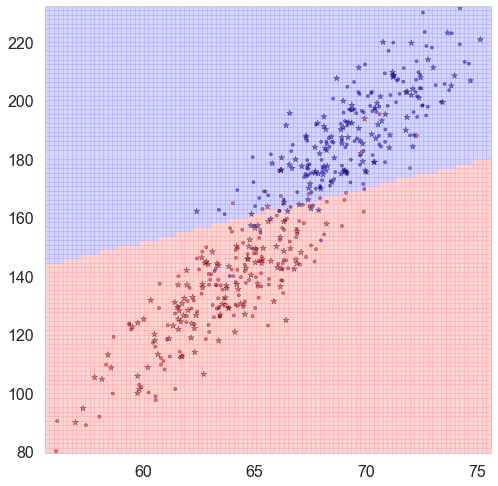
Binary Classification results based on two features
cmap_light = ListedColormap(['#FFAAAA', '#AAFFAA', '#AAAAFF'])
cmap_bold = ListedColormap(['#FF0000', '#00FF00', '#0000FF'])
def classify_plot(ax, Xtr, Xte, ytr, yte, clf, mesh=True, colorscale=cmap_light, cdiscrete=cmap_bold, alpha=0.1, psize=10):
h = .02
X=np.concatenate((Xtr, Xte))
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
xx, yy = np.meshgrid(np.linspace(x_min, x_max, 100),
np.linspace(y_min, y_max, 100))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
ZZ = Z.reshape(xx.shape)
if mesh:
plt.pcolormesh(xx, yy, ZZ, cmap = colorscale, alpha=alpha, axes=ax)
else:
showtr = ytr
showte = yte
ax.scatter(Xtr.iloc[:, 0], Xtr.iloc[:, 1], c=showtr-1, cmap=cdiscrete, s=psize, alpha=alpha, edgecolor="k")
# and testing points
ax.scatter(Xte.iloc[:, 0], Xte.iloc[:, 1], c=showte-1, cmap=cdiscrete, alpha=alpha, marker="*", s=psize+30)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
return ax,xx,yy
Parameters
-
ax: figure information -
Xtr: pandas.DataFrameTraining set of features where features are in the columns
-
Xte: DataFrameTesting set of features where features are in the columns
-
ytr: DataFrameTraining set of target
-
yte: DataFrameTesting set of target
-
clf: fitted classifier -
mesh: plot mesh or not. IfFalse, then it only plots predicted values. -
colorscale: specify theColormapfor mesh plotColormapis the base classes to convert numbers to color to the RGBA color. -
cdiscrete: specify theColormapfor points -
alpha: transparency -
psize: size of points
Returns
-
ax:figure information -
xx: x axis coordinates -
yy: y axis coordinates
Demo:
dfhw=pd.read_csv("https://dl.dropboxusercontent.com/u/75194/stats/data/01_heights_weights_genders.csv")
df=dfhw.sample(500, replace=False)
df.Gender = (df.Gender=="Male") * 1
X_train, X_test, y_train, y_test = train_test_split(
df[['Height', 'Weight']], df.Gender, test_size=0.4, random_state=42)
clflog = LogisticRegression()
clf = clflog.fit(X_train, y_train)
plt.figure(figsize = (15,13))
ax=plt.gca()
classify_plot(ax, X_train, X_test, y_train, y_test, clf, alpha = 0.5)
plt.show()
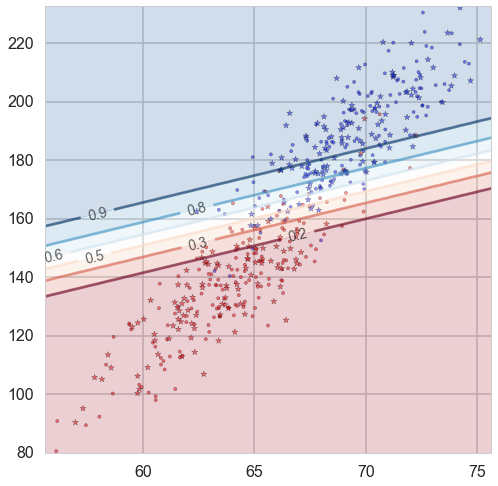
Probability boundaries
cm = plt.cm.RdBu
def classify_plot_prob(ax, Xtr, Xte, ytr, yte, clf, colorscale=cmap_light, cdiscrete=cmap_bold, ccolor=cm, psize=10, alpha=0.1, prob=True):
ax,xx,yy = points_plot(ax, Xtr, Xte, ytr, yte, clf, mesh=False, colorscale=colorscale, cdiscrete=cdiscrete, psize=psize, alpha=alpha)
if prob:
Z = clf.predict_proba(np.c_[xx.ravel(), yy.ravel()])[:, 1]
else:
Z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contourf(xx, yy, Z, cmap=ccolor, alpha=.2, axes=ax)
cs2 = plt.contour(xx, yy, Z, cmap=ccolor, alpha=.6, axes=ax)
plt.clabel(cs2, fmt = '%2.1f', colors = 'k', fontsize=14, axes=ax)
return ax
Parameters
-
ccolor: specify theColormapfor contour plot -
prob: use probability or decision function
Demo:
plt.figure(figsize = (15,13))
ax=plt.gca()
classify_plot_prob(ax, X_train, X_test, y_train, y_test, clf, alpha = 0.5)
plt.show()
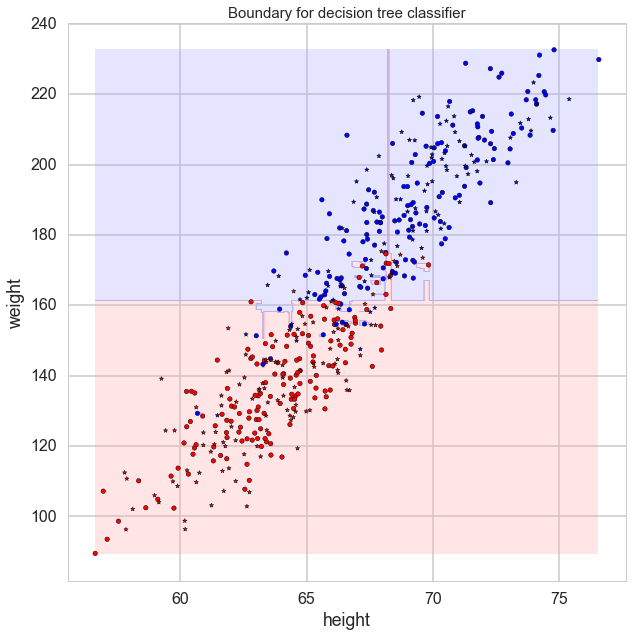
Tree Classifier plot
def plot_tree(ax, Xtr, Xte, ytr, yte, clf, plot_train = True, plot_test = True, lab = ['Feature 1', 'Feature 2'],
mesh=True, colorscale=cmap_light, cdiscrete=cmap_bold, alpha=0.3, psize=10):
# Create a meshgrid as our test data
plt.figure(figsize=(15,10))
plot_step= 0.05
xmin, xmax= Xtr[:,0].min(), Xtr[:,0].max()
ymin, ymax= Xtr[:,1].min(), Xtr[:,1].max()
xx, yy = np.meshgrid(np.arange(xmin, xmax, plot_step), np.arange(ymin, ymax, plot_step) )
# Re-cast every coordinate in the meshgrid as a 2D point
Xplot= np.c_[xx.ravel(), yy.ravel()]
# Predict the class
Z = clf.predict( Xplot )
# Re-shape the results
Z= Z.reshape( xx.shape )
cs = ax.contourf(xx, yy, Z, cmap= cmap_light, alpha=0.3)
# Overlay training samples
if (plot_train == True):
ax.scatter(Xtr[:, 0], Xtr[:, 1], c=ytr-1, cmap=cmap_bold, alpha=alpha,edgecolor="k")
# and testing points
if (plot_test == True):
ax.scatter(Xte[:, 0], Xte[:, 1], c=yte-1, cmap=cmap_bold, alpha=alpha, marker="*")
ax.set_xlabel(lab[0])
ax.set_ylabel(lab[1])
ax.set_title("Boundary for decision tree classifier",fontsize=15)
Demo
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(X_train, y_train)
plt.figure(figsize = (10,10))
ax=plt.gca()
plot_tree(ax, np.array(X_train), np.array(X_test), np.array(y_train), np.array(y_test),
clf = rf, lab = ['height', 'weight'], alpha = 1)
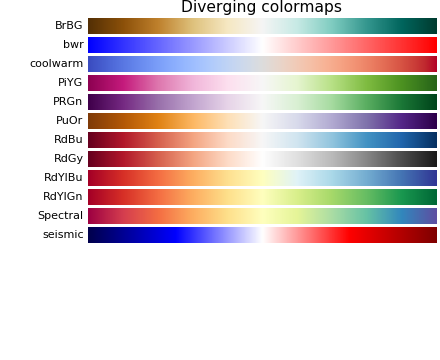
Colormaps
Colormaps is very useful when we try to convert numbers into colors.
If the colormaps have a listed values like ListedColormap(['#FFAAAA', '#AAFFAA', '#AAAAFF']), it means to map one number into one of these three colors. For example if \(X = [x_1, … , x_n]\) has values from 0 to 1, then [0, 0.33] indicates color #FFAAAA, (0.33, 0.66] indicates color #AAFFAA and (0.66, 1] indicates color AAAAFF.
If the colormaps is continuous like plt.cm.RdBu, then the numbers should map to that range. The following is one example:
Precision vs. Recall
def pr_curve(truthvec, scorevec, digit_prec=2):
threshvec = np.unique(np.round(scorevec,digit_prec))
numthresh = len(threshvec)
tpvec = np.zeros(numthresh)
fpvec = np.zeros(numthresh)
fnvec = np.zeros(numthresh)
for i in range(numthresh):
thresh = threshvec[i]
tpvec[i] = sum(truthvec[scorevec>=thresh])
fpvec[i] = sum(1-truthvec[scorevec>=thresh])
fnvec[i] = sum(truthvec[scorevec<thresh])
recallvec = tpvec/(tpvec + fnvec)
precisionvec = tpvec/(tpvec + fpvec)
plt.plot(precisionvec,recallvec)
plt.axis([0, 1, 0, 1])
plt.xlabel('precision')
plt.ylabel('recall')
return (recallvec, precisionvec, threshvec)
Demo
rf = RandomForestClassifier()
rf.fit(X_train, y_train)
y_pred = rf.predict(X_test)
plt.figure(figsize = (10,10))
pr_curve(y_test, y_pred)
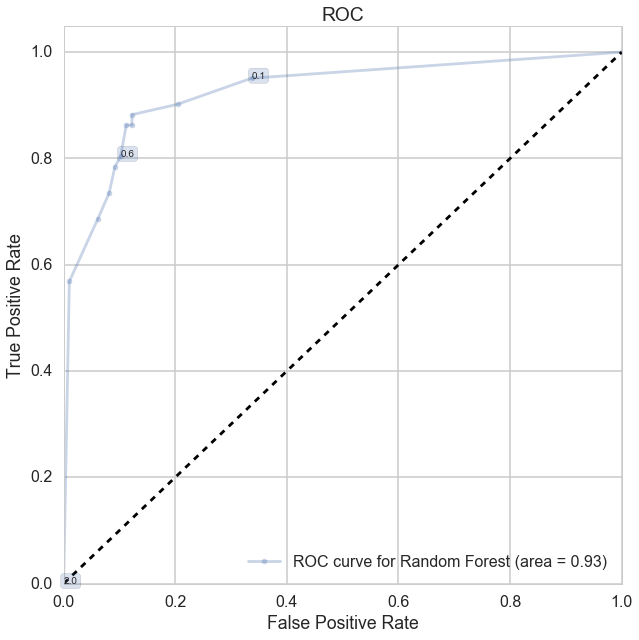
ROC Curve
This function can present the thresholds in the ROC figure.
def make_roc(name, clf, ytest, xtest, ax=None, labe=5, proba=True, skip=0):
initial=False
if not ax:
ax=plt.gca()
initial=True
if proba:
fpr, tpr, thresholds=roc_curve(ytest, clf.predict_proba(xtest)[:,1])
else:
fpr, tpr, thresholds=roc_curve(ytest, clf.decision_function(xtest))
roc_auc = auc(fpr, tpr)
if skip:
l=fpr.shape[0]
ax.plot(fpr[0:l:skip], tpr[0:l:skip], '.-', alpha=0.3, label='ROC curve for %s (area = %0.2f)' % (name, roc_auc))
else:
ax.plot(fpr, tpr, '.-', alpha=0.3, label='ROC curve for %s (area = %0.2f)' % (name, roc_auc))
label_kwargs = {}
label_kwargs['bbox'] = dict(
boxstyle='round,pad=0.3', alpha=0.2,
)
for k in range(0, fpr.shape[0],labe):
#from https://gist.github.com/podshumok/c1d1c9394335d86255b8
threshold = str(np.round(thresholds[k], 2))
ax.annotate(threshold, (fpr[k], tpr[k]), **label_kwargs)
if initial:
ax.plot([0, 1], [0, 1], 'k--')
ax.set_xlim([0.0, 1.0])
ax.set_ylim([0.0, 1.05])
ax.set_xlabel('False Positive Rate')
ax.set_ylabel('True Positive Rate')
ax.set_title('ROC')
ax.legend(loc="lower right")
print('AUC: ', roc_auc)
return ax
Parameters
-
name: string, indicating the name of model -
clf: fitted classifier -
labe: the distance between two adjacent labels. -
proba: some classifiers, like KNN, Logistic regression, need to setproba = True -
skip: the step size when plot ROC
Demo
plt.figure(figsize = (10,10))
make_roc('Random Forest', rf, y_test, X_test)
plt.show()
Remarks
- The first two functions are only for logistic regression, KNN, SVM and Random Forest.
- When you try to feed SVM into
classify_plot_prob, you need to setprobability = TrueinSVC(). - The second function may present ugly figures for KNN and Random Forest.